Covid-19-visualizations
COVID-19 Iowa
Ian Lyttle 2020-09-02
library("fs")
library("glue")
library("tidyverse")
## ── Attaching packages ────────────────────────────────────────────────────────────────────────── tidyverse 1.3.0 ──
## ✓ ggplot2 3.3.2 ✓ purrr 0.3.4
## ✓ tibble 3.0.3 ✓ dplyr 1.0.1
## ✓ tidyr 1.1.1 ✓ stringr 1.4.0
## ✓ readr 1.3.1.9000 ✓ forcats 0.5.0
## ── Conflicts ───────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
## x dplyr::collapse() masks glue::collapse()
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
library("USAboundaries") # also install_github("ropensci/USAboundariesData")
library("sf")
## Linking to GEOS 3.8.1, GDAL 3.1.1, PROJ 6.3.1
library("colorspace")
dir_source <- path("data", "wrangle")
dir_target <- path("data", "iowa")
dir_create(dir_target)
Prepare data
Let’s read in the county data:
iowa_counties <-
read_csv(path(dir_source, "iowa-counties.csv")) %>%
print()
## Parsed with column specification:
## cols(
## date = col_date(format = ""),
## county = col_character(),
## cases = col_double(),
## deaths = col_double(),
## new_cases = col_double(),
## new_deaths = col_double(),
## new_cases_week_avg = col_double(),
## new_deaths_week_avg = col_double(),
## aggregation = col_character()
## )
## # A tibble: 15,145 x 9
## date county cases deaths new_cases new_deaths new_cases_week_…
## <date> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020-09-02 Polk 13511 226 46 0 203.
## 2 2020-09-02 Woodb… 4157 56 5 0 21.1
## 3 2020-09-02 Johns… 4098 26 47 0 165.
## 4 2020-09-02 Black… 3830 76 0 0 35.7
## 5 2020-09-02 Linn 3005 94 12 0 25
## 6 2020-09-02 Story 2549 16 15 0 97
## 7 2020-09-02 Dallas 2391 38 5 0 31.9
## 8 2020-09-02 Scott 2254 21 5 0 27.6
## 9 2020-09-02 Dubuq… 2062 36 13 0 17.7
## 10 2020-09-02 Buena… 1828 12 1 0 0.857
## # … with 15,135 more rows, and 2 more variables: new_deaths_week_avg <dbl>,
## # aggregation <chr>
Let’s have a quick look at all the counties.
ggplot(iowa_counties, aes(date, cases)) +
geom_line(aes(color = county)) +
scale_y_log10()
This plot is not all that informative, except to tell us that we have the data we expect.
Let’s get just the data for the most-current data.
iowa_counties_current <-
iowa_counties %>%
filter(date == max(date)) %>%
arrange(desc(cases)) %>%
print()
## # A tibble: 99 x 9
## date county cases deaths new_cases new_deaths new_cases_week_…
## <date> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020-09-02 Polk 13511 226 46 0 203.
## 2 2020-09-02 Woodb… 4157 56 5 0 21.1
## 3 2020-09-02 Johns… 4098 26 47 0 165.
## 4 2020-09-02 Black… 3830 76 0 0 35.7
## 5 2020-09-02 Linn 3005 94 12 0 25
## 6 2020-09-02 Story 2549 16 15 0 97
## 7 2020-09-02 Dallas 2391 38 5 0 31.9
## 8 2020-09-02 Scott 2254 21 5 0 27.6
## 9 2020-09-02 Dubuq… 2062 36 13 0 17.7
## 10 2020-09-02 Buena… 1828 12 1 0 0.857
## # … with 89 more rows, and 2 more variables: new_deaths_week_avg <dbl>,
## # aggregation <chr>
It could be useful to see the current distribution of number of cases per county:
date_max <- unique(iowa_counties_current$date)
ggplot(iowa_counties_current) +
geom_histogram(aes(x = cases), binwidth = 2) +
theme_bw() +
labs(
x = "number of reported cases in county",
y = "number of counties",
title = glue("Iowa: distribution of counties by number of COVID-19 cases - {date_max}"),
caption = params$nyt_citation
)
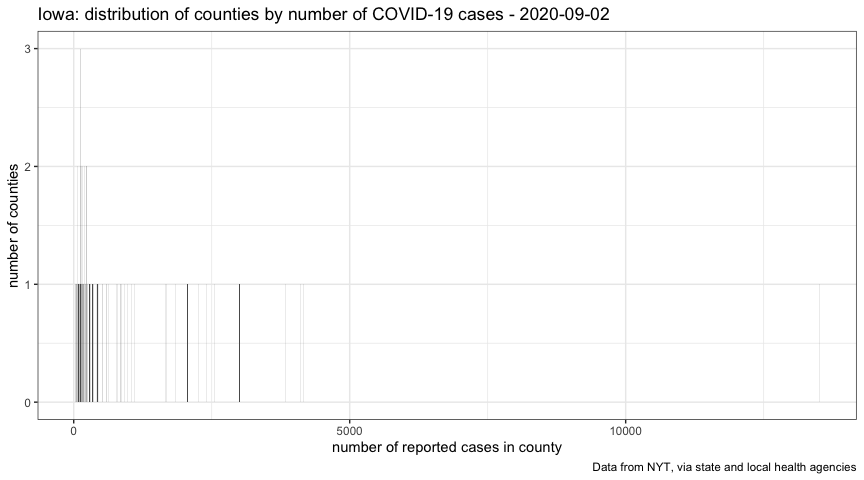
On the right, we see Johnson, Polk, and Linn counties, with the greatest number of reported cases. On the left, we see a lot of counties with only a few cases.
Our eyes will let us pay attention to only a finite number of things at a time, so I propose to set a threshold of 50 cases. We identify those counties where the number of cases is greater than or equal to the threshold.
counties_large <-
iowa_counties_current %>%
filter(cases >= params$threshold_cases) %>%
arrange(desc(cases)) %>%
pull(county)
counties_large
## [1] "Polk" "Woodbury" "Johnson" "Black Hawk"
## [5] "Linn" "Story" "Dallas" "Scott"
## [9] "Dubuque" "Buena Vista" "Pottawattamie" "Marshall"
## [13] "Wapello" "Webster" "Muscatine" "Sioux"
## [17] "Clinton" "Cerro Gordo" "Crawford" "Warren"
## [21] "Plymouth" "Tama" "Jasper" "Des Moines"
## [25] "Wright" "Marion" "Lee" "Dickinson"
## [29] "Carroll" "Louisa" "Boone" "Washington"
## [33] "Bremer" "Franklin" "Henry" "Hamilton"
## [37] "Clarke" "Clay" "Delaware" "Hardin"
## [41] "Winneshiek" "Mahaska" "Emmet" "Floyd"
## [45] "Jackson" "Shelby" "Butler" "Benton"
## [49] "O'Brien" "Poweshiek" "Clayton" "Allamakee"
## [53] "Buchanan" "Jones" "Guthrie" "Madison"
## [57] "Winnebago" "Hancock" "Cedar" "Lyon"
## [61] "Humboldt" "Harrison" "Fayette" "Cherokee"
## [65] "Howard" "Grundy" "Mills" "Pocahontas"
## [69] "Calhoun" "Kossuth" "Sac" "Jefferson"
## [73] "Palo Alto" "Iowa" "Monroe" "Mitchell"
## [77] "Page" "Taylor" "Chickasaw" "Cass"
## [81] "Monona" "Van Buren" "Osceola" "Lucas"
## [85] "Union" "Appanoose" "Davis" "Montgomery"
## [89] "Worth" "Keokuk" "Fremont" "Greene"
## [93] "Wayne"
In addition to compiliing the data for the counties with large numbers of cases, we also create aggregeted datasets that show:
- the total for the entire state.
- the total for all the counties that are not considered individually.
iowa_total <-
iowa_counties %>%
group_by(date) %>%
summarize(
county = factor(NA_character_, levels = counties_large),
cases = sum(cases),
deaths = sum(deaths),
new_cases = sum(new_cases),
new_deaths = sum(new_deaths),
new_cases_week_avg = sum(new_cases_week_avg),
new_deaths_week_avg = sum(new_deaths_week_avg),
aggregation = "total"
) %>%
arrange(desc(date)) %>%
print()
## `summarise()` ungrouping output (override with `.groups` argument)
## # A tibble: 179 x 9
## date county cases deaths new_cases new_deaths new_cases_week_…
## <date> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020-09-02 <NA> 66136 1125 295 0 1042.
## 2 2020-09-01 <NA> 65841 1125 607 5 1178.
## 3 2020-08-31 <NA> 65234 1120 668 8 1188.
## 4 2020-08-30 <NA> 64566 1112 747 3 1162.
## 5 2020-08-29 <NA> 63819 1109 1010 3 1120.
## 6 2020-08-28 <NA> 62809 1106 1337 15 1070
## 7 2020-08-27 <NA> 61472 1091 2627 14 993.
## 8 2020-08-26 <NA> 58845 1077 1252 18 758.
## 9 2020-08-25 <NA> 57593 1059 677 13 676.
## 10 2020-08-24 <NA> 56916 1046 487 10 631.
## # … with 169 more rows, and 2 more variables: new_deaths_week_avg <dbl>,
## # aggregation <chr>
iowa_remainder <-
iowa_counties %>%
filter(!(county %in% counties_large)) %>%
group_by(date) %>%
summarize(
county = factor(NA_character_, levels = counties_large),
cases = sum(cases),
deaths = sum(deaths),
new_cases = sum(new_cases),
new_deaths = sum(new_deaths),
new_cases_week_avg = sum(new_cases_week_avg),
new_deaths_week_avg = sum(new_deaths_week_avg),
aggregation = "remainder"
) %>%
arrange(desc(date)) %>%
print()
## `summarise()` ungrouping output (override with `.groups` argument)
## # A tibble: 170 x 9
## date county cases deaths new_cases new_deaths new_cases_week_…
## <date> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020-09-02 <NA> 232 3 2 0 3.86
## 2 2020-09-01 <NA> 230 3 3 0 3.86
## 3 2020-08-31 <NA> 227 3 3 0 3.71
## 4 2020-08-30 <NA> 224 3 3 0 4
## 5 2020-08-29 <NA> 221 3 2 0 3.86
## 6 2020-08-28 <NA> 219 3 3 0 4.29
## 7 2020-08-27 <NA> 216 3 11 0 4.29
## 8 2020-08-26 <NA> 205 3 2 0 3.57
## 9 2020-08-25 <NA> 203 3 2 0 3.43
## 10 2020-08-24 <NA> 201 3 5 0 3.57
## # … with 160 more rows, and 2 more variables: new_deaths_week_avg <dbl>,
## # aggregation <chr>
iowa_counties_large <-
iowa_counties %>%
filter(county %in% counties_large) %>%
mutate(
county = factor(county, levels = counties_large)
)
Let’s combine our datasets:
data_combined <- bind_rows(iowa_counties_large, iowa_total, iowa_remainder)
If we wanted do one of those FT-style charts, we need to choose an index day for each county and aggregation. We choose an arbitrary threshold of 20 reported cases.
data_combined_index <-
data_combined %>%
group_by(aggregation, county) %>%
arrange(date) %>%
mutate(
exceeds_threshold_cases = cases >= params$threshold_index_cases,
index_cases = cumsum(exceeds_threshold_cases) - 1,
exceeds_threshold_new_cases = new_cases >= params$threshold_new_cases,
index_new_cases = cumsum(cumany(exceeds_threshold_new_cases)) - 1
)
We combine all these datasets into a plot:
plot_cases <-
ggplot(data_combined, aes(date, cases)) +
geom_line(aes(color = county, linetype = aggregation)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = county),
show.legend = FALSE
) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_linetype(
name = "grouping",
breaks = c("total", "remainder", "none"),
labels = c("state total", "all other counties", "county")
) +
scale_color_discrete_qualitative(
breaks = counties_large,
palette = params$cat_palette,
na.value = "grey50"
) +
coord_cartesian(
ylim = c(10, NA)
) +
theme_bw() +
labs(
x = NULL,
y = "number of reported cases",
title = "Iowa: COVID-19 cases by reporting date",
subtitle = "Includes counties with most reported cases, also aggregations",
caption = params$nyt_citation
)
plot_new_cases_week_avg <-
ggplot(data_combined, aes(date, new_cases_week_avg + 0.01)) +
geom_line(aes(color = county, linetype = aggregation)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = county),
show.legend = FALSE
) +
scale_y_log10(
labels = function(x) {round(x, 1)},
sec.axis = dup_axis(name = NULL)
) +
scale_linetype(
name = "grouping",
breaks = c("total", "remainder", "none"),
labels = c("state total", "all other counties", "county")
) +
scale_color_discrete_qualitative(
breaks = counties_large,
palette = params$cat_palette,
na.value = "grey50"
) +
theme_bw() +
labs(
x = NULL,
y = "number of new reported cases",
title = "Iowa: COVID-19 daily new cases (7-day rolling average) by reporting date",
subtitle = "Includes counties with most reported cases, also aggregations",
caption = params$nyt_citation
)
# function that takes a set of limits, returns the breaks
# - we want breaks every 7 days.
f_break <- function(limits) {
low <- floor(limits[1] / 7) * 7
hi <- ceiling(limits[2] / 7) * 7
seq(low, hi, by = 7)
}
# function that returns data for "doubling" lines
#' @param data `data.frame`
#' @param var_index `character`, name of variable in `data` containing day-index
#' @param var_number `character`, name of variable in `data` contiaining number-data
#' @param days_doubling `numeric`, vector with number-of-days to include
#'
#' @return `data.frame` with variables `days_doubling`, `index`, `number`, `label`
#'
f_doubling <- function(data, var_index, var_number, threshold_number,
days_doubling = c(2, 4, 7, 14)) {
index_range <- range(data[[var_index]])
number_range <- range(data[[var_number]])
max_number <- number_range[2]
max_index <- index_range[2]
# for each `days_doubling`, determine the index when days are maximum
by_number <-
crossing(days_doubling, number = max_number) %>%
mutate(
index = days_doubling * log(number/threshold_number, 2)
)
by_index <-
crossing(days_doubling, index = index_range) %>%
mutate(
number = threshold_number * 2^(index/days_doubling)
)
combined <-
dplyr::bind_rows(by_index, by_number) %>%
dplyr::select(days_doubling, index, number) %>%
dplyr::mutate(
label = ifelse(index > 0, glue::glue("{days_doubling}-day doubling"), "")
) %>%
dplyr::filter(index <= max_index, number <= max_number) %>%
dplyr::arrange(days_doubling, index)
combined
}
plot_cases_ft <-
ggplot(
data_combined_index %>% filter(index_cases >= 0),
aes(index_cases, cases)
) +
geom_line(
data =
. %>%
f_doubling("index_cases", "cases", params$threshold_index_cases),
aes(x = index, y = number, group = factor(days_doubling)),
color = "grey80",
linetype = "twodash"
) +
geom_text(
data =
. %>%
f_doubling("index_cases", "cases", params$threshold_index_cases) %>%
filter(str_length(label) > 0),
aes(x = index, y = number, group = factor(days_doubling), label = label),
hjust = "right",
nudge_y = 0.05,
color = "grey80"
) +
geom_line(
aes(color = county, linetype = aggregation),
alpha = 0.25
) +
geom_line(
data = . %>% filter(date >= max(date) - 14),
aes(color = county, linetype = aggregation),
alpha = 0.5
) +
geom_line(
data = . %>% filter(date >= max(date) - 7),
aes(color = county, linetype = aggregation),
alpha = 1
) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = county),
show.legend = FALSE
) +
geom_text(
data = . %>% filter(date == max(date), aggregation == "none"),
aes(x = index_cases, label = county, color = county),
hjust = "left",
nudge_x = .5,
size = 3,
show.legend = FALSE
) +
geom_text(
data = . %>%
ungroup() %>%
filter(date == max(date), aggregation != "none") %>%
mutate(
label = fct_recode(
aggregation,
`state total` = "total",
`all other counties` = "remainder"
)
),
aes(x = index_cases, label = label),
color = "grey10",
hjust = "left",
nudge_x = 0.5,
size = 3,
show.legend = FALSE
) +
scale_x_continuous(breaks = f_break, expand = expansion(mult = c(0, 0.1))) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_linetype(
name = "grouping",
breaks = c("total", "remainder", "none"),
labels = c("state total", "all other counties", "county")
) +
scale_color_discrete_qualitative(
breaks = counties_large,
palette = params$cat_palette,
na.value = "grey50"
) +
guides(color = FALSE, linetype = FALSE) +
theme_bw() +
labs(
x = glue("days since {params$threshold_index_cases} cases"),
y = "number of reported cases",
title = glue("Iowa: COVID-19 cumulative-case trajectories as of {max(data_combined_index$date)}"),
subtitle = "Includes counties with most reported cases, also aggregations",
caption = params$nyt_citation
)
plot_new_cases_week_avg_ft <-
ggplot(
data_combined_index %>% filter(index_new_cases >= 0),
aes(index_new_cases, new_cases_week_avg + 0.01)
) +
geom_line(
aes(color = county, linetype = aggregation),
alpha = 0.25
) +
geom_line(
data = . %>% filter(date >= max(date) - 14),
aes(color = county, linetype = aggregation),
alpha = 0.5
) +
geom_line(
data = . %>% filter(date >= max(date) - 7),
aes(color = county, linetype = aggregation),
alpha = 1
) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = county),
show.legend = FALSE
) +
geom_text(
data = . %>% filter(date == max(date), aggregation == "none"),
aes(x = index_new_cases, label = county, color = county),
hjust = "left",
nudge_x = .5,
size = 3,
show.legend = FALSE
) +
geom_text(
data = . %>%
ungroup() %>%
filter(date == max(date), aggregation != "none") %>%
mutate(
label = fct_recode(
aggregation,
NULL = "county",
`state total` = "total",
`all other counties` = "remainder"
)
),
aes(x = index_new_cases, label = label),
color = "grey10",
hjust = "left",
nudge_x = 0.5,
size = 3,
show.legend = FALSE
) +
scale_x_continuous(breaks = f_break, expand = expansion(mult = c(0, 0.1))) +
scale_y_log10(
labels = function(x) {round(x, 1)},
sec.axis = dup_axis(name = NULL)
) +
scale_linetype(
name = "grouping",
breaks = c("total", "remainder", "none"),
labels = c("state total", "all other counties", "county")
) +
scale_color_discrete_qualitative(
breaks = counties_large,
palette = params$cat_palette,
na.value = "grey50"
) +
guides(color = FALSE, linetype = FALSE) +
theme_bw() +
labs(
x = glue("days since {params$threshold_index_new_cases} new cases"),
y = "number of new reported cases",
title = glue("Iowa: COVID-19 new-case (7-day rolling average) trajectories as of {max(data_combined_index$date)}"),
subtitle = "Includes counties with most reported cases, also aggregations",
caption = params$nyt_citation
)
County groups
Let’s look at the poulation of counties. I’d like to sort Iowa’s counties into four groups, ordered by county-population, such that each group of counties contains approximately the same population.
iowa_county_population <-
read_csv(path(dir_source, "iowa-county-population.csv")) %>%
print()
## Parsed with column specification:
## cols(
## fips = col_double(),
## county = col_character(),
## population = col_double(),
## cumulative_poulation = col_double(),
## quartile_population = col_double(),
## population_group = col_character()
## )
## # A tibble: 99 x 6
## fips county population cumulative_poula… quartile_popula… population_group
## <dbl> <chr> <dbl> <dbl> <dbl> <chr>
## 1 19153 Polk 490161 3155070 1 large
## 2 19113 Linn 226706 2664909 0.845 large
## 3 19163 Scott 172943 2438203 0.773 mid-large
## 4 19103 Johnson 151140 2265260 0.718 mid-large
## 5 19013 Black H… 131228 2114120 0.670 mid-large
## 6 19193 Woodbury 103107 1982892 0.628 mid-large
## 7 19061 Dubuque 97311 1879785 0.596 mid-large
## 8 19169 Story 97117 1782474 0.565 mid-large
## 9 19049 Dallas 93453 1685357 0.534 mid-large
## 10 19155 Pottawa… 93206 1591904 0.505 mid-large
## # … with 89 more rows
You can see that the “first” group contains two counties: Polk and Linn; the “second” group contains seven counties, from Scott to Dallas. This second group of counties is home to the Regents’ Universities, Iowa’s part of the Quad cities, Dubuque, Sioux City, and suburban Des Moines.
The “third” and “fourth” groups contain 24 and 66 counties, respectively. Here’s a map of Iowa’s counties, showing these groups:
iowa_map <- us_counties(resolution = "low", state = "Iowa")
I am interested to compare how the trajectory of recorded cases behaves, or might behave differently, among these groups of counties.
group_colors <-
colorspace::sequential_hcl(palette = "Blues 3", n = 6)[1:4] # drop lightest
iowa_map_with_group <-
iowa_map %>%
left_join(
iowa_county_population %>% select(county, population_group),
by = c(name = "county")
)
plot_iowa_map_group <-
ggplot() +
geom_sf(
data = iowa_map_with_group,
aes(fill = population_group),
color = "white"
) +
scale_fill_manual(values = group_colors) +
labs(
title = "Iowa counties by population-group",
subtitle = "Each population-group has about 1/4 of state's population (2019 estimate)",
caption = params$iowa_citation,
fill = "group"
) +
theme_minimal() +
theme(
panel.grid.major = element_blank(),
axis.text = element_blank()
)
Let’s look at proportions of counties, population, and (most-recent) reported cases and deaths.
iowa_counts <-
iowa_county_population %>% select(county, population, population_group) %>%
left_join(
iowa_counties_current,
by = "county"
) %>%
mutate(
cases = ifelse(is.na(cases), 0, cases),
deaths = ifelse(is.na(deaths), 0, deaths)
) %>%
group_by(population_group) %>%
summarize(
counties = n(),
population = sum(population),
cases = sum(cases),
deaths = sum(deaths)
) %>%
print()
## `summarise()` ungrouping output (override with `.groups` argument)
## # A tibble: 4 x 5
## population_group counties population cases deaths
## <chr> <int> <dbl> <dbl> <dbl>
## 1 large 2 716867 16516 320
## 2 mid-large 8 939505 23005 303
## 3 mid-small 23 719312 13815 296
## 4 small 66 779386 12800 206
vars_count <- c("counties", "population", "cases", "deaths")
iowa_counts_tall <-
iowa_counts %>%
pivot_longer(
all_of(vars_count),
names_to = "category",
values_to = "count"
) %>%
mutate(
category = factor(category, rev(vars_count))
) %>%
print()
## # A tibble: 16 x 3
## population_group category count
## <chr> <fct> <dbl>
## 1 large counties 2
## 2 large population 716867
## 3 large cases 16516
## 4 large deaths 320
## 5 mid-large counties 8
## 6 mid-large population 939505
## 7 mid-large cases 23005
## 8 mid-large deaths 303
## 9 mid-small counties 23
## 10 mid-small population 719312
## 11 mid-small cases 13815
## 12 mid-small deaths 296
## 13 small counties 66
## 14 small population 779386
## 15 small cases 12800
## 16 small deaths 206
plot_iowa_proportions <-
ggplot(
iowa_counts_tall,
aes(x = category, y = count)
) +
geom_col(aes(fill = population_group), position = position_fill(reverse = TRUE)) +
scale_y_continuous(labels = scales::percent) +
scale_fill_manual(values = group_colors) +
coord_flip() +
labs(
x = "",
y = "proportion",
fill = "group",
title = glue("Iowa: COVID-19 proportions - as of {date_max}"),
caption = params$nyt_citation
) +
theme_bw()
iowa_counties_group <-
iowa_counties %>%
left_join(
iowa_county_population %>% select(county, population_group),
by = "county"
)
iowa_counties_group_agg <-
iowa_counties_group %>%
group_by(date, population_group) %>%
summarize(
cases = sum(cases),
deaths = sum(deaths),
new_cases = sum(new_cases),
new_deaths = sum(new_deaths),
new_cases_week_avg = sum(new_cases_week_avg),
new_deaths_week_avg = sum(new_deaths_week_avg)
) %>%
ungroup()
## `summarise()` regrouping output by 'date' (override with `.groups` argument)
plot_cases_group <-
ggplot(iowa_counties_group_agg, aes(date, cases)) +
geom_line(
aes(color = population_group)
) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = population_group),
show.legend = FALSE
) +
scale_color_manual(values = group_colors) +
scale_y_log10(sec.axis = dup_axis(name = NULL)) +
theme_bw() +
labs(
x = NULL,
y = "number of reported cases",
color = "group",
title = "Iowa: COVID-19 cases by reporting date",
caption = params$nyt_citation
)
plot_new_cases_group <-
ggplot(iowa_counties_group_agg, aes(date, new_cases_week_avg)) +
geom_line(
aes(color = population_group)
) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = population_group),
show.legend = FALSE
) +
scale_color_manual(values = group_colors) +
scale_y_log10(sec.axis = dup_axis(name = NULL)) +
theme_bw() +
labs(
x = NULL,
y = "number of new reported cases",
color = "group",
title = "Iowa: COVID-19 daily new cases (7-day rolling average) by reporting date",
caption = params$nyt_citation
)
iowa_counties_group_agg_ft <-
iowa_counties_group_agg %>%
group_by(population_group) %>%
arrange(date) %>%
mutate(
exceeds_threshold_cases = cases > params$threshold_index_cases,
index_cases = cumsum(exceeds_threshold_cases) - 1,
exceeds_threshold_new_cases = new_cases >= params$threshold_index_new_cases,
index_new_cases = cumsum(cumany(exceeds_threshold_new_cases)) - 1
)
plot_cases_group_ft <-
ggplot(
iowa_counties_group_agg_ft %>% filter(exceeds_threshold_cases),
aes(index_cases, cases)) +
geom_line(
data =
. %>%
f_doubling("index_cases", "cases", params$threshold_index_cases),
aes(x = index, y = number, group = factor(days_doubling)),
color = "grey80",
linetype = "twodash"
) +
geom_text(
data =
. %>%
f_doubling("index_cases", "cases", params$threshold_index_cases) %>%
filter(str_length(label) > 0),
aes(x = index, y = number, label = label),
hjust = "right",
nudge_y = 0.05,
color = "grey80"
) +
geom_line(aes(color = population_group)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = population_group),
show.legend = FALSE
) +
scale_x_continuous(breaks = f_break) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_color_manual(values = group_colors) +
theme_bw() +
labs(
x = glue("days since {params$threshold_index_cases} cases"),
y = "number of reported cases",
color = "group",
title = "Iowa: COVID-19 cumulative-case trajectories",
caption = params$nyt_citation
)
plot_new_cases_group_ft <-
ggplot(
iowa_counties_group_agg_ft %>% filter(index_new_cases >= 0),
aes(index_new_cases, new_cases_week_avg)) +
geom_line(aes(color = population_group)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = population_group),
show.legend = FALSE
) +
scale_x_continuous(breaks = f_break) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_color_manual(values = group_colors) +
theme_bw() +
labs(
x = glue("days since {params$threshold_index_new_cases} new cases"),
y = "number of new reported cases",
color = "group",
title = "Iowa: COVID-19 new-case trajectories (7-day rolling average)",
caption = params$nyt_citation
)
plot_iowa_deaths_ft <-
ggplot(
data_combined_index %>% filter(aggregation == "total", deaths > 0),
aes(x = date, y = deaths)
) +
geom_line() +
geom_point(
data = . %>% filter(date == max(date))
) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
theme_bw() +
labs(
x = NULL,
y = "total number of fatalities",
title = "Iowa: COVID-19 fatality trajectory",
caption = params$nyt_citation
)
counties_closed_may_01 <-
c(
"Allamakee",
"Benton",
"Black Hawk",
"Bremer",
"Dallas",
"Des Moines",
"Dubuque",
"Fayette",
"Henry",
"Iowa",
"Jasper",
"Johnson",
"Linn",
"Louisa",
"Marshall",
"Muscatine",
"Polk",
"Poweshiek",
"Scott",
"Tama",
"Washington",
"Woodbury"
)
iowa_reopen <-
iowa_counties %>%
mutate(
open_may_01 = !(county %in% counties_closed_may_01)
) %>%
group_by(date, open_may_01) %>%
summarize(
cases = sum(cases),
deaths = sum(deaths),
new_cases = sum(new_cases),
new_deaths = sum(new_deaths),
new_cases_week_avg = sum(new_cases_week_avg),
new_deaths_week_avg = sum(new_deaths_week_avg)
) %>%
ungroup()
## `summarise()` regrouping output by 'date' (override with `.groups` argument)
iowa_reopen_annotate <-
tibble(
date = as.Date(c("2020-04-24", "2020-05-01", "2020-05-15")),
event = c("announced", "initial reopening", "later reopening")
)
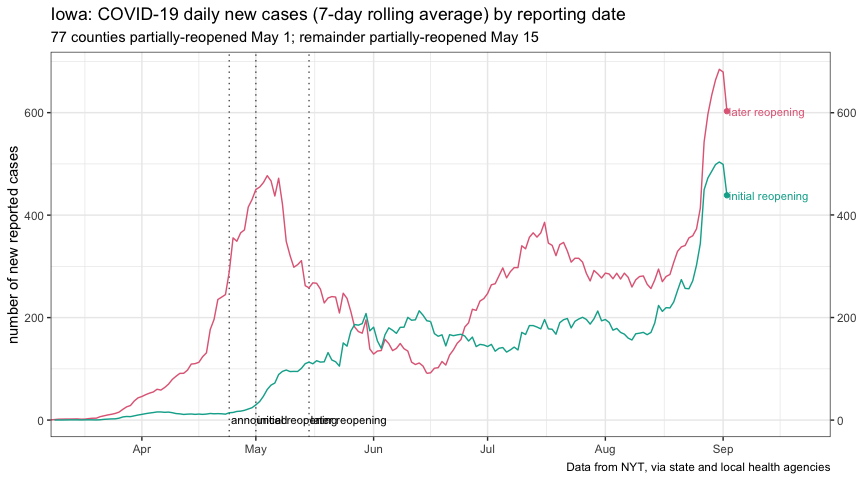
plot_reopen_cases <-
ggplot(iowa_reopen, aes(date, cases)) +
geom_vline(
data = iowa_reopen_annotate,
aes(xintercept = date),
linetype = "dotted",
alpha = 0.7
) +
geom_text(
data = iowa_reopen_annotate,
aes(x = date, label = event),
y = 1,
size = 3,
hjust = "left",
nudge_x = 0.5
) +
geom_line(aes(color = open_may_01)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = open_may_01)
) +
geom_text(
data =
. %>%
filter(date == max(date)) %>%
mutate(label = ifelse(open_may_01, "initial reopening", "later reopening")),
aes(label = label, color = open_may_01),
hjust = "left",
nudge_x = .5,
size = 3,
show.legend = FALSE
) +
scale_x_date(expand = expansion(mult = c(0, 0.15))) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_color_discrete_qualitative(palette = params$cat_palette) +
guides(color = FALSE) +
coord_cartesian(
ylim = c(1, NA)
) +
theme_bw() +
labs(
x = NULL,
y = "number of reported cases",
title = "Iowa: COVID-19 cumulative-case trajectories",
subtitle = "77 counties partially-reopened May 1; remainder partially-reopened May 15",
caption = params$nyt_citation
)
plot_reopen_new_cases <-
ggplot(iowa_reopen, aes(date, new_cases_week_avg)) +
geom_vline(
data = iowa_reopen_annotate,
aes(xintercept = date),
linetype = "dotted",
alpha = 0.7
) +
geom_text(
data = iowa_reopen_annotate,
aes(x = date, label = event),
y = 1,
size = 3,
hjust = "left",
nudge_x = 0.5
) +
geom_line(aes(color = open_may_01)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = open_may_01)
) +
geom_text(
data =
. %>%
filter(date == max(date)) %>%
mutate(label = ifelse(open_may_01, "initial reopening", "later reopening")),
aes(label = label, color = open_may_01),
hjust = "left",
nudge_x = .5,
size = 3,
show.legend = FALSE
) +
scale_x_date(expand = expansion(mult = c(0, 0.15))) +
scale_y_log10(
sec.axis = dup_axis(name = NULL)
) +
scale_color_discrete_qualitative(palette = params$cat_palette) +
guides(color = FALSE) +
coord_cartesian(
ylim = c(1, NA)
) +
theme_bw() +
labs(
x = NULL,
y = "number of new reported cases",
title = "Iowa: COVID-19 daily new cases (7-day rolling average) by reporting date",
subtitle = "77 counties partially-reopened May 1; remainder partially-reopened May 15",
caption = params$nyt_citation
)
plot_reopen_new_cases_linear <-
plot_reopen_new_cases +
scale_y_continuous(
sec.axis = dup_axis(name = NULL)
)
## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
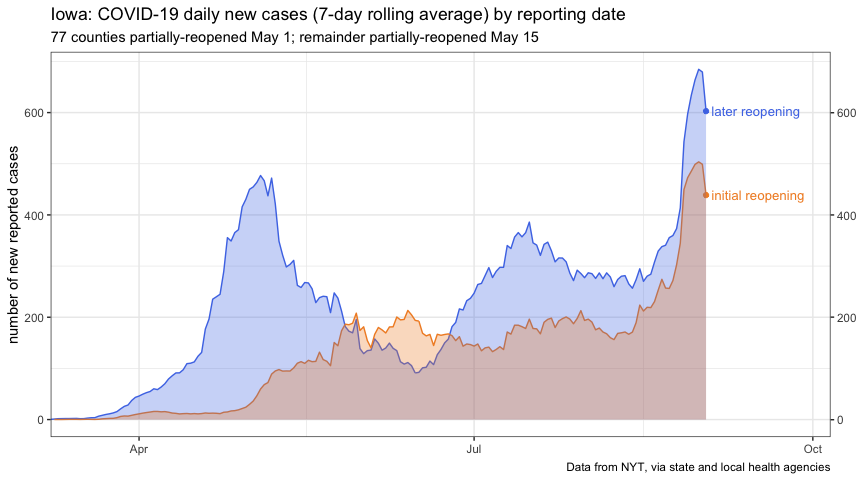
Let’s imagine an area-chart:
iowa_reopen_pyramid <-
iowa_reopen %>%
mutate(
across(
.cols = matches("cases|deaths"),
~ ifelse(open_may_01, -.x, .x)
)
)
scale_color <- c(`TRUE` = "#F28E2B", `FALSE` = "#4E79E7")
ggplot(
iowa_reopen,
aes(x = date, y = new_cases_week_avg, group = open_may_01)
) +
geom_line(aes(color = open_may_01)) +
geom_point(
data = . %>% filter(date == max(date)),
aes(color = open_may_01)
) +
geom_area(
aes(fill = open_may_01),
alpha = 0.3,
position = "identity",
) +
geom_text(
data =
. %>%
filter(date == max(date)) %>%
mutate(
label = ifelse(open_may_01, "initial reopening", "later reopening")
),
aes(label = label, color = open_may_01),
hjust = "left",
nudge_x = 1.5,
size = 3.5,
show.legend = FALSE
) +
scale_x_date(expand = expansion(mult = c(0, 0.18))) +
scale_y_continuous(
sec.axis = dup_axis(name = NULL),
labels = abs
) +
scale_color_manual(values = scale_color) +
scale_fill_manual(values = scale_color) +
guides(color = FALSE, fill = FALSE) +
theme_bw() +
labs(
x = NULL,
y = "number of new reported cases",
title = "Iowa: COVID-19 daily new cases (7-day rolling average) by reporting date",
subtitle = "77 counties partially-reopened May 1; remainder partially-reopened May 15",
caption = params$nyt_citation
)
Discussion
plot_cases
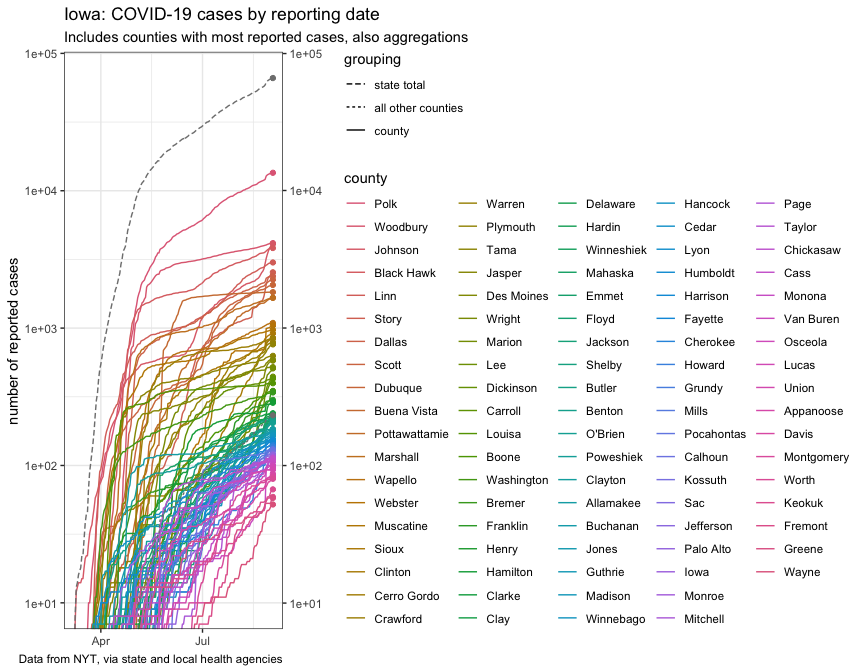
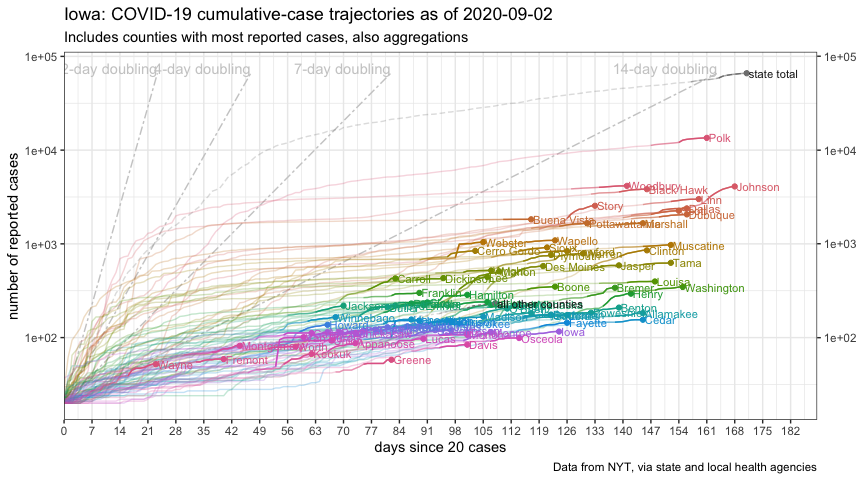
For another view, here’s an FT-style chart:
plot_cases_ft
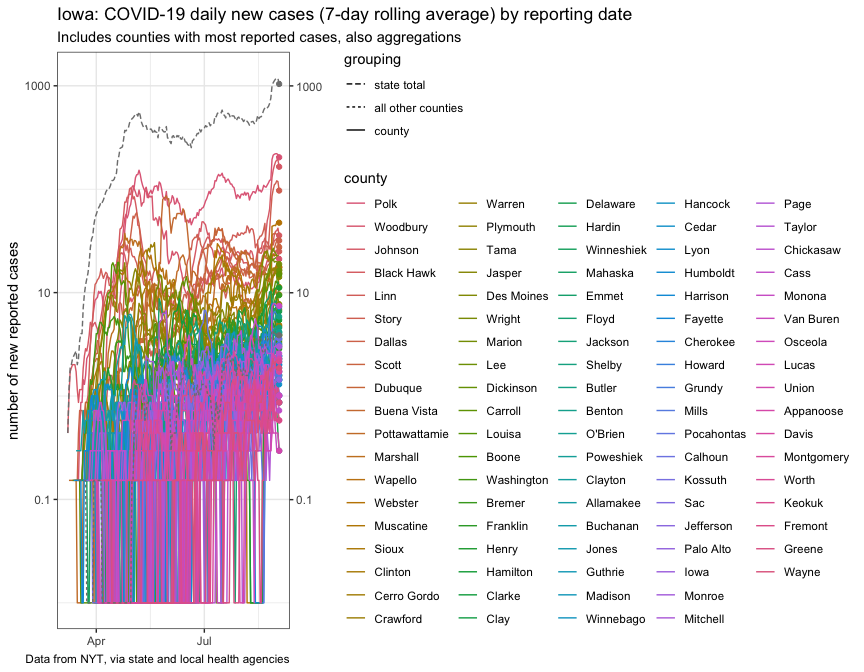
plot_new_cases_week_avg
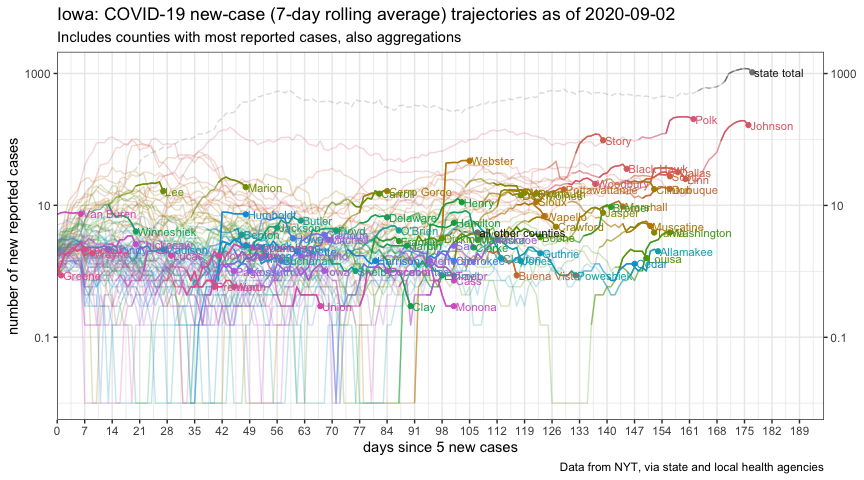
plot_new_cases_week_avg_ft
plot_iowa_map_group
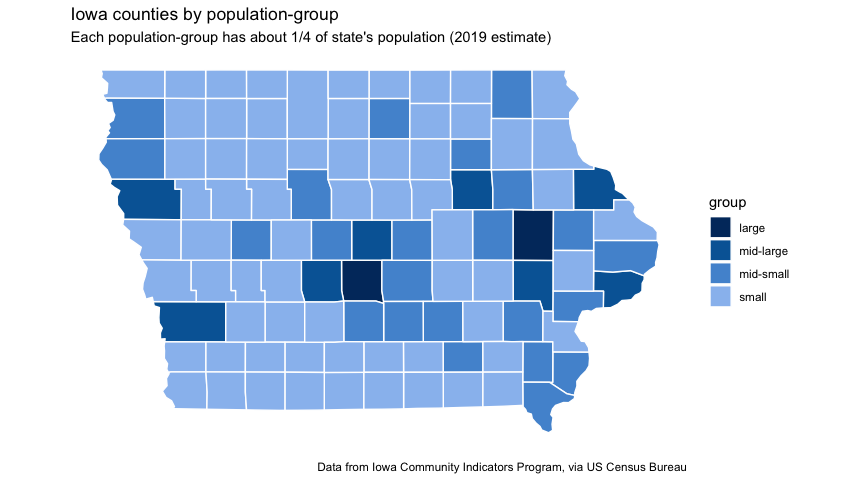
plot_iowa_proportions
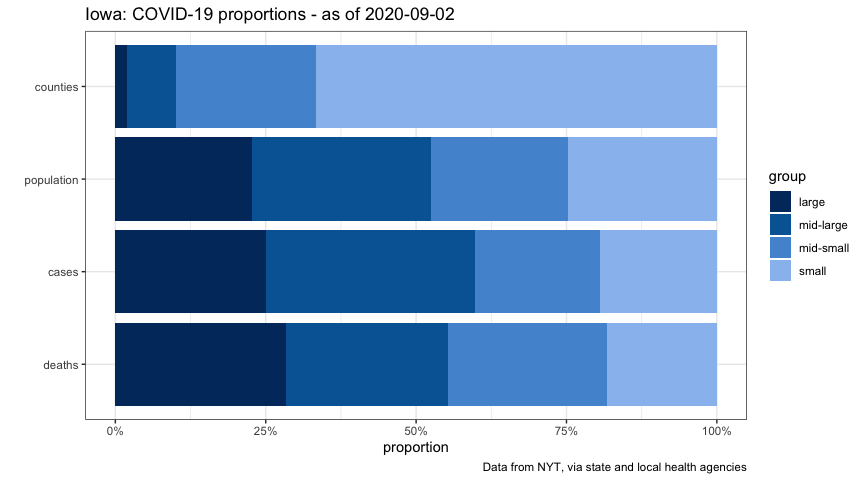
plot_cases_group
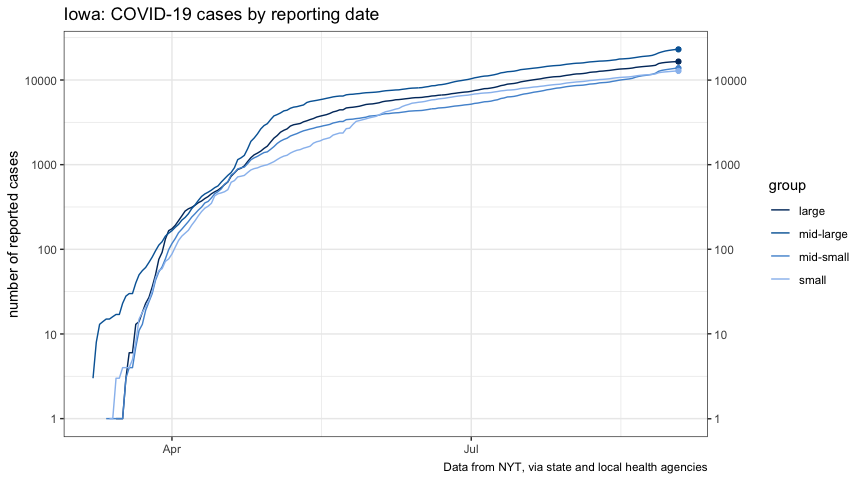
plot_new_cases_group
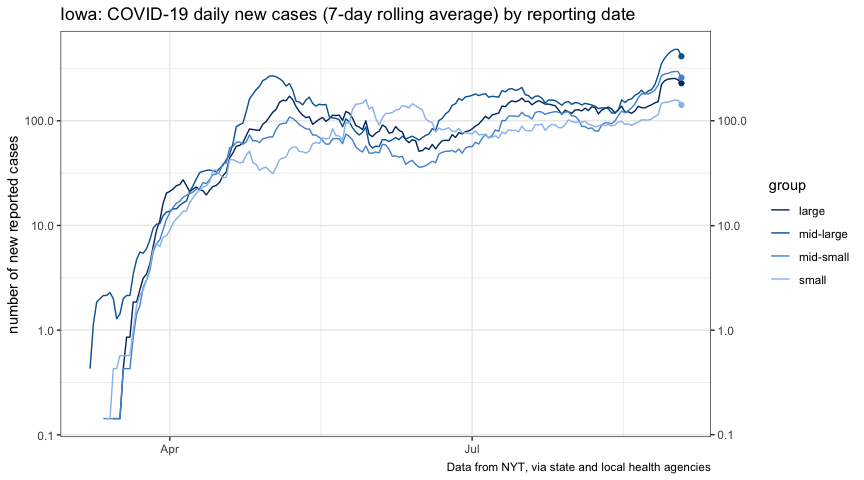
plot_cases_group_ft
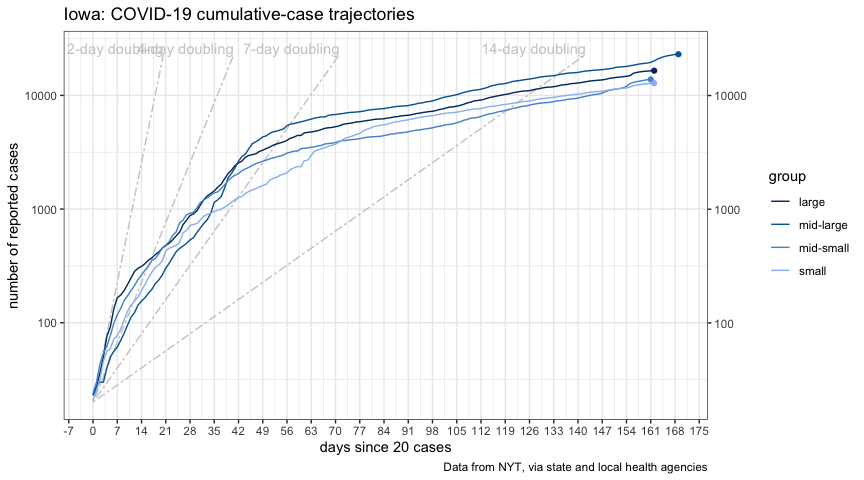
plot_new_cases_group_ft
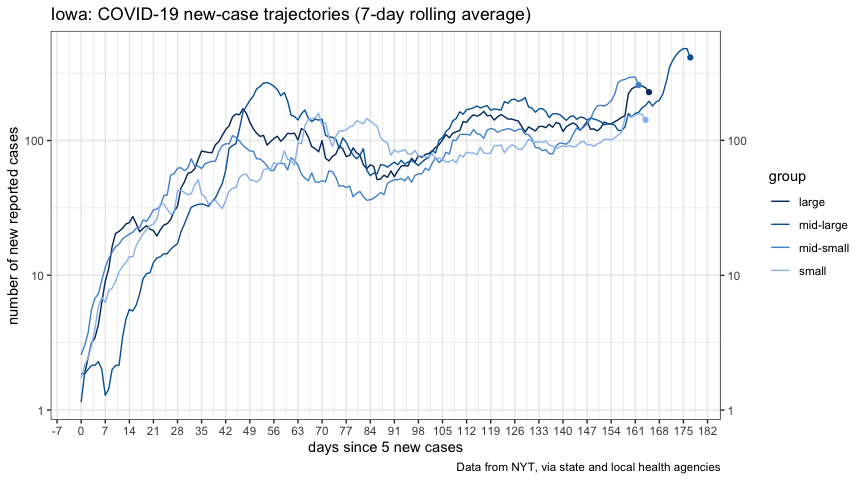
plot_iowa_deaths_ft
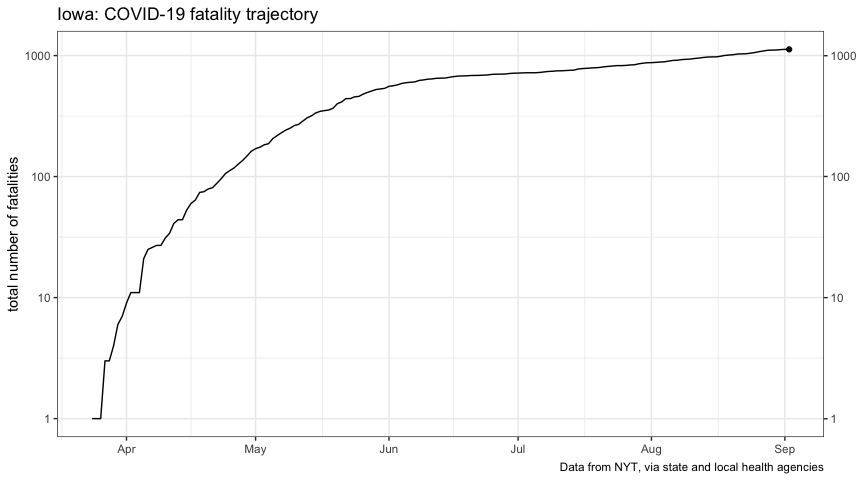
plot_reopen_cases
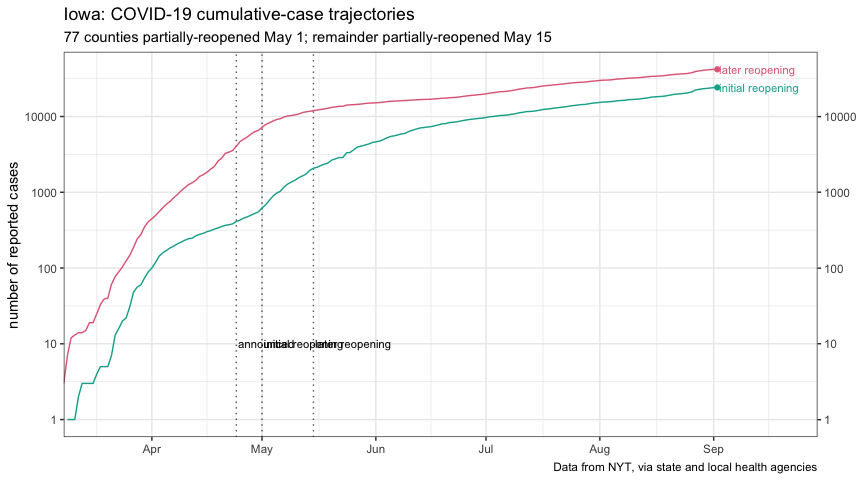
plot_reopen_new_cases
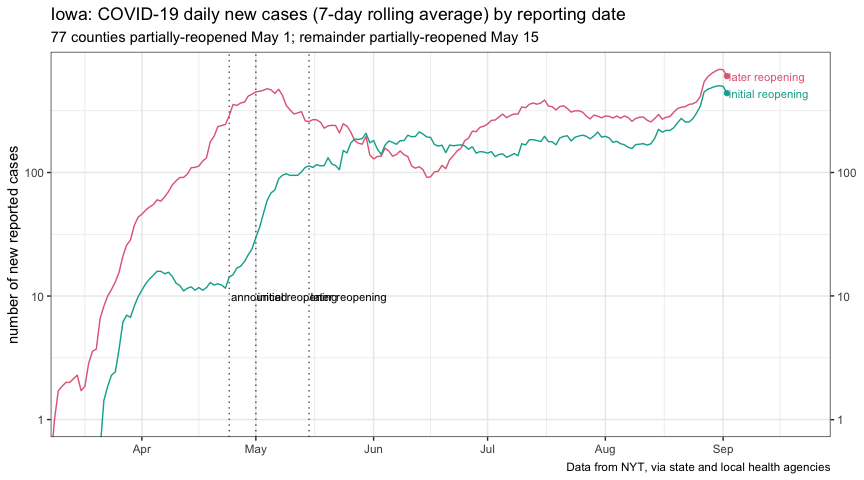
plot_reopen_new_cases_linear